pyiron#
Define workflow with pyiron_base#
import numpy as np
from pyiron_base import job
from python_workflow_definition.pyiron_base import write_workflow_json
from workflow import (
calculate_qe as _calculate_qe,
generate_structures as _generate_structures,
get_bulk_structure as _get_bulk_structure,
plot_energy_volume_curve as _plot_energy_volume_curve,
)
workflow_json_filename = "pyiron_base_qe.json"
calculate_qe = job(_calculate_qe, output_key_lst=["energy", "volume", "structure"])
generate_structures = job(_generate_structures)
plot_energy_volume_curve = job(_plot_energy_volume_curve)
get_bulk_structure = job(_get_bulk_structure)
pseudopotentials = {"Al": "Al.pbe-n-kjpaw_psl.1.0.0.UPF"}
structure = get_bulk_structure(
element="Al",
a=4.04,
cubic=True,
)
calc_mini = calculate_qe(
working_directory="mini",
input_dict={
"structure": structure,
"pseudopotentials": pseudopotentials,
"kpts": (3, 3, 3),
"calculation": "vc-relax",
"smearing": 0.02,
},
)
number_of_strains = 5
structure_lst = generate_structures( # the generate_structures() function is not available in the workflow graph
structure=calc_mini.output.structure,
strain_lst=np.linspace(0.9, 1.1, number_of_strains),
list_length=number_of_strains,
)
job_strain_lst = []
for i, structure_strain in enumerate(structure_lst):
calc_strain = calculate_qe(
working_directory="strain_" + str(i),
input_dict={
"structure": structure_strain,
"pseudopotentials": pseudopotentials,
"kpts": (3, 3, 3),
"calculation": "scf",
"smearing": 0.02,
},
)
job_strain_lst.append(calc_strain)
plot = plot_energy_volume_curve(
volume_lst=[job.output.volume for job in job_strain_lst],
energy_lst=[job.output.energy for job in job_strain_lst],
)
write_workflow_json(delayed_object=plot, file_name=workflow_json_filename)
!cat {workflow_json_filename}
{
"version": "0.1.0",
"nodes": [
{
"id": 0,
"type": "function",
"value": "workflow.plot_energy_volume_curve"
},
{
"id": 1,
"type": "function",
"value": "python_workflow_definition.shared.get_list"
},
{
"id": 2,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 3,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 4,
"type": "function",
"value": "workflow.generate_structures"
},
{
"id": 5,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 6,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 7,
"type": "function",
"value": "workflow.get_bulk_structure"
},
{
"id": 8,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 9,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 10,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 11,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 12,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 13,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 14,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 15,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 16,
"type": "function",
"value": "python_workflow_definition.shared.get_list"
},
{
"id": 17,
"type": "input",
"name": "working_directory_0",
"value": "strain_0"
},
{
"id": 18,
"type": "input",
"name": "working_directory_1",
"value": "mini"
},
{
"id": 19,
"type": "input",
"name": "element",
"value": "Al"
},
{
"id": 20,
"type": "input",
"name": "a",
"value": 4.04
},
{
"id": 21,
"type": "input",
"name": "cubic",
"value": true
},
{
"id": 22,
"type": "input",
"name": "pseudopotentials",
"value": {
"Al": "Al.pbe-n-kjpaw_psl.1.0.0.UPF"
}
},
{
"id": 23,
"type": "input",
"name": "kpts",
"value": [
3,
3,
3
]
},
{
"id": 24,
"type": "input",
"name": "calculation_0",
"value": "vc-relax"
},
{
"id": 25,
"type": "input",
"name": "smearing",
"value": 0.02
},
{
"id": 26,
"type": "input",
"name": "strain_lst",
"value": [
0.9,
0.9500000000000001,
1.0,
1.05,
1.1
]
},
{
"id": 27,
"type": "input",
"name": "calculation_1",
"value": "scf"
},
{
"id": 28,
"type": "input",
"name": "working_directory_2",
"value": "strain_1"
},
{
"id": 29,
"type": "input",
"name": "working_directory_3",
"value": "strain_2"
},
{
"id": 30,
"type": "input",
"name": "working_directory_4",
"value": "strain_3"
},
{
"id": 31,
"type": "input",
"name": "working_directory_5",
"value": "strain_4"
},
{
"id": 32,
"type": "output",
"name": "result"
}
],
"edges": [
{
"target": 0,
"targetPort": "volume_lst",
"source": 1,
"sourcePort": null
},
{
"target": 1,
"targetPort": "0",
"source": 2,
"sourcePort": "volume"
},
{
"target": 2,
"targetPort": "working_directory",
"source": 17,
"sourcePort": null
},
{
"target": 2,
"targetPort": "input_dict",
"source": 3,
"sourcePort": null
},
{
"target": 3,
"targetPort": "structure",
"source": 4,
"sourcePort": "s_0"
},
{
"target": 4,
"targetPort": "structure",
"source": 5,
"sourcePort": "structure"
},
{
"target": 5,
"targetPort": "working_directory",
"source": 18,
"sourcePort": null
},
{
"target": 5,
"targetPort": "input_dict",
"source": 6,
"sourcePort": null
},
{
"target": 6,
"targetPort": "structure",
"source": 7,
"sourcePort": null
},
{
"target": 7,
"targetPort": "element",
"source": 19,
"sourcePort": null
},
{
"target": 7,
"targetPort": "a",
"source": 20,
"sourcePort": null
},
{
"target": 7,
"targetPort": "cubic",
"source": 21,
"sourcePort": null
},
{
"target": 6,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 6,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 6,
"targetPort": "calculation",
"source": 24,
"sourcePort": null
},
{
"target": 6,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 4,
"targetPort": "strain_lst",
"source": 26,
"sourcePort": null
},
{
"target": 3,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 3,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 3,
"targetPort": "calculation",
"source": 27,
"sourcePort": null
},
{
"target": 3,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 1,
"targetPort": "1",
"source": 8,
"sourcePort": "volume"
},
{
"target": 8,
"targetPort": "working_directory",
"source": 28,
"sourcePort": null
},
{
"target": 8,
"targetPort": "input_dict",
"source": 9,
"sourcePort": null
},
{
"target": 9,
"targetPort": "structure",
"source": 4,
"sourcePort": "s_1"
},
{
"target": 9,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 9,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 9,
"targetPort": "calculation",
"source": 27,
"sourcePort": null
},
{
"target": 9,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 1,
"targetPort": "2",
"source": 10,
"sourcePort": "volume"
},
{
"target": 10,
"targetPort": "working_directory",
"source": 29,
"sourcePort": null
},
{
"target": 10,
"targetPort": "input_dict",
"source": 11,
"sourcePort": null
},
{
"target": 11,
"targetPort": "structure",
"source": 4,
"sourcePort": "s_2"
},
{
"target": 11,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 11,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 11,
"targetPort": "calculation",
"source": 27,
"sourcePort": null
},
{
"target": 11,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 1,
"targetPort": "3",
"source": 12,
"sourcePort": "volume"
},
{
"target": 12,
"targetPort": "working_directory",
"source": 30,
"sourcePort": null
},
{
"target": 12,
"targetPort": "input_dict",
"source": 13,
"sourcePort": null
},
{
"target": 13,
"targetPort": "structure",
"source": 4,
"sourcePort": "s_3"
},
{
"target": 13,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 13,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 13,
"targetPort": "calculation",
"source": 27,
"sourcePort": null
},
{
"target": 13,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 1,
"targetPort": "4",
"source": 14,
"sourcePort": "volume"
},
{
"target": 14,
"targetPort": "working_directory",
"source": 31,
"sourcePort": null
},
{
"target": 14,
"targetPort": "input_dict",
"source": 15,
"sourcePort": null
},
{
"target": 15,
"targetPort": "structure",
"source": 4,
"sourcePort": "s_4"
},
{
"target": 15,
"targetPort": "pseudopotentials",
"source": 22,
"sourcePort": null
},
{
"target": 15,
"targetPort": "kpts",
"source": 23,
"sourcePort": null
},
{
"target": 15,
"targetPort": "calculation",
"source": 27,
"sourcePort": null
},
{
"target": 15,
"targetPort": "smearing",
"source": 25,
"sourcePort": null
},
{
"target": 0,
"targetPort": "energy_lst",
"source": 16,
"sourcePort": null
},
{
"target": 16,
"targetPort": "0",
"source": 2,
"sourcePort": "energy"
},
{
"target": 16,
"targetPort": "1",
"source": 8,
"sourcePort": "energy"
},
{
"target": 16,
"targetPort": "2",
"source": 10,
"sourcePort": "energy"
},
{
"target": 16,
"targetPort": "3",
"source": 12,
"sourcePort": "energy"
},
{
"target": 16,
"targetPort": "4",
"source": 14,
"sourcePort": "energy"
},
{
"target": 32,
"targetPort": null,
"source": 0,
"sourcePort": null
}
]
}
Load Workflow with aiida#
from aiida import orm, load_profile
load_profile()
Profile<uuid='1682a1dbda304ec998030372ef5350ab' name='pwd'>
from python_workflow_definition.aiida import load_workflow_json
wg = load_workflow_json(file_name=workflow_json_filename)
wg.nodes.get_bulk_structure.inputs.a.value = orm.Float(4.05)
wg.run()
05/26/2025 04:46:41 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_bulk_structure8
05/26/2025 04:46:41 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_bulk_structure8, type: PyFunction, finished.
05/26/2025 04:46:41 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict7
05/26/2025 04:46:42 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict7, type: PyFunction, finished.
05/26/2025 04:46:42 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe6
[jupyter-pythonworkflow-fl--x---218119f8:01776] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:47:34 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['energy', 'volume'])
05/26/2025 04:47:34 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe6, type: PyFunction, finished.
05/26/2025 04:47:34 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: generate_structures5
05/26/2025 04:47:35 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: generate_structures5, type: PyFunction, finished.
05/26/2025 04:47:35 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict4,get_dict10,get_dict12,get_dict14,get_dict16
05/26/2025 04:47:36 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict4, type: PyFunction, finished.
05/26/2025 04:47:36 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe3,get_dict10,get_dict12,get_dict14,get_dict16
[jupyter-pythonworkflow-fl--x---218119f8:01828] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:47:47 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 04:47:47 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe3, type: PyFunction, finished.
05/26/2025 04:47:47 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict10,get_dict12,get_dict14,get_dict16
05/26/2025 04:47:48 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict10, type: PyFunction, finished.
05/26/2025 04:47:48 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe9,get_dict12,get_dict14,get_dict16
[jupyter-pythonworkflow-fl--x---218119f8:01860] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:47:59 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 04:48:00 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe9, type: PyFunction, finished.
05/26/2025 04:48:00 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict12,get_dict14,get_dict16
05/26/2025 04:48:00 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict12, type: PyFunction, finished.
05/26/2025 04:48:00 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe11,get_dict14,get_dict16
[jupyter-pythonworkflow-fl--x---218119f8:01874] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:48:13 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 04:48:13 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe11, type: PyFunction, finished.
05/26/2025 04:48:13 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict14,get_dict16
05/26/2025 04:48:14 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict14, type: PyFunction, finished.
05/26/2025 04:48:14 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe13,get_dict16
[jupyter-pythonworkflow-fl--x---218119f8:01885] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:48:29 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 04:48:29 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe13, type: PyFunction, finished.
05/26/2025 04:48:29 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict16
05/26/2025 04:48:29 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_dict16, type: PyFunction, finished.
05/26/2025 04:48:29 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe15
[jupyter-pythonworkflow-fl--x---218119f8:01895] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:48:44 AM <1688> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 04:48:44 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: calculate_qe15, type: PyFunction, finished.
05/26/2025 04:48:44 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_list2,get_list17
05/26/2025 04:48:44 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_list2, type: PyFunction, finished.
05/26/2025 04:48:44 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_list17
05/26/2025 04:48:45 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: get_list17, type: PyFunction, finished.
05/26/2025 04:48:45 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run: plot_energy_volume_curve1
05/26/2025 04:48:45 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|update_task_state]: Task: plot_energy_volume_curve1, type: PyFunction, finished.
05/26/2025 04:48:46 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|continue_workgraph]: tasks ready to run:
05/26/2025 04:48:46 AM <1688> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [17|WorkGraphEngine|finalize]: Finalize workgraph.
Load Workflow with jobflow#
from python_workflow_definition.jobflow import load_workflow_json
from jobflow.managers.local import run_locally
flow = load_workflow_json(file_name=workflow_json_filename)
flow[0].function_kwargs["a"] = 4.05
result = run_locally(flow)
result
2025-05-26 04:48:47,564 INFO Started executing jobs locally
2025-05-26 04:48:47,751 INFO Starting job - get_bulk_structure (f386f375-fcc9-4700-a077-b8c7b4ed29d4)
2025-05-26 04:48:47,754 INFO Finished job - get_bulk_structure (f386f375-fcc9-4700-a077-b8c7b4ed29d4)
2025-05-26 04:48:47,755 INFO Starting job - get_dict (6c1c9b0f-5c14-4ae9-be98-984400228394)
2025-05-26 04:48:47,757 INFO Finished job - get_dict (6c1c9b0f-5c14-4ae9-be98-984400228394)
2025-05-26 04:48:47,757 INFO Starting job - calculate_qe (f511b680-fa97-420f-904d-2ae232dbe75b)
[jupyter-pythonworkflow-fl--x---218119f8:01905] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
2025-05-26 04:49:37,980 INFO Finished job - calculate_qe (f511b680-fa97-420f-904d-2ae232dbe75b)
2025-05-26 04:49:37,981 INFO Starting job - generate_structures (d5fc7015-e7fe-4111-bf16-23bdc6755f93)
2025-05-26 04:49:37,985 INFO Finished job - generate_structures (d5fc7015-e7fe-4111-bf16-23bdc6755f93)
2025-05-26 04:49:37,986 INFO Starting job - get_dict (49b7b2e8-78ca-4d63-bfa6-f3b47ca3378f)
2025-05-26 04:49:37,988 INFO Finished job - get_dict (49b7b2e8-78ca-4d63-bfa6-f3b47ca3378f)
2025-05-26 04:49:37,988 INFO Starting job - get_dict (a45e0be5-c229-4817-8dc8-e2f54c46205f)
2025-05-26 04:49:37,990 INFO Finished job - get_dict (a45e0be5-c229-4817-8dc8-e2f54c46205f)
2025-05-26 04:49:37,990 INFO Starting job - get_dict (30bf90f1-57c2-474b-8bd2-d8a18061d85c)
2025-05-26 04:49:37,992 INFO Finished job - get_dict (30bf90f1-57c2-474b-8bd2-d8a18061d85c)
2025-05-26 04:49:37,993 INFO Starting job - get_dict (d4b595f2-93a9-42d7-a547-f64be9b4ca80)
2025-05-26 04:49:37,995 INFO Finished job - get_dict (d4b595f2-93a9-42d7-a547-f64be9b4ca80)
2025-05-26 04:49:37,996 INFO Starting job - get_dict (8c32a293-38d8-4c3a-82fc-78edd324d64d)
2025-05-26 04:49:37,998 INFO Finished job - get_dict (8c32a293-38d8-4c3a-82fc-78edd324d64d)
2025-05-26 04:49:37,998 INFO Starting job - calculate_qe (e747fb6a-e704-44ff-906e-664be75da6fc)
[jupyter-pythonworkflow-fl--x---218119f8:01923] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
2025-05-26 04:49:48,629 INFO Finished job - calculate_qe (e747fb6a-e704-44ff-906e-664be75da6fc)
2025-05-26 04:49:48,630 INFO Starting job - calculate_qe (579fbf80-e6e7-464c-a808-f5cecd9ef430)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:01934] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
2025-05-26 04:50:00,439 INFO Finished job - calculate_qe (579fbf80-e6e7-464c-a808-f5cecd9ef430)
2025-05-26 04:50:00,439 INFO Starting job - calculate_qe (ceb70e3f-c90a-4aa3-a670-23ecf40fa11b)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:01945] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
2025-05-26 04:50:13,181 INFO Finished job - calculate_qe (ceb70e3f-c90a-4aa3-a670-23ecf40fa11b)
2025-05-26 04:50:13,182 INFO Starting job - calculate_qe (290a2a1d-9a74-40b1-af28-e4f61b357694)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:01955] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
2025-05-26 04:50:27,249 INFO Finished job - calculate_qe (290a2a1d-9a74-40b1-af28-e4f61b357694)
2025-05-26 04:50:27,250 INFO Starting job - calculate_qe (a9f810e8-c291-46b9-ab8f-4c59765bf3b9)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:01965] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
2025-05-26 04:50:37,525 INFO Finished job - calculate_qe (a9f810e8-c291-46b9-ab8f-4c59765bf3b9)
2025-05-26 04:50:37,526 INFO Starting job - get_list (20122c29-0e2d-4a9c-8506-d371390f8042)
2025-05-26 04:50:37,528 INFO Finished job - get_list (20122c29-0e2d-4a9c-8506-d371390f8042)
2025-05-26 04:50:37,529 INFO Starting job - get_list (9842e2f0-3ca3-4a23-b82b-a3644a0294a8)
2025-05-26 04:50:37,531 INFO Finished job - get_list (9842e2f0-3ca3-4a23-b82b-a3644a0294a8)
2025-05-26 04:50:37,532 INFO Starting job - plot_energy_volume_curve (a1614162-0427-4cd2-a2c4-01837c5ae505)
2025-05-26 04:50:37,581 INFO Finished job - plot_energy_volume_curve (a1614162-0427-4cd2-a2c4-01837c5ae505)
2025-05-26 04:50:37,582 INFO Finished executing jobs locally
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
{'f386f375-fcc9-4700-a077-b8c7b4ed29d4': {1: Response(output='{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.05, 0.0, 0.0], [0.0, 4.05, 0.0], [0.0, 0.0, 4.05]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.025, 2.025], [2.025, 0.0, 2.025], [2.025, 2.025, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'6c1c9b0f-5c14-4ae9-be98-984400228394': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.05, 0.0, 0.0], [0.0, 4.05, 0.0], [0.0, 0.0, 4.05]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.025, 2.025], [2.025, 0.0, 2.025], [2.025, 2.025, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'vc-relax', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'f511b680-fa97-420f-904d-2ae232dbe75b': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.045637215946841, 0.0, 0.0], [0.0, 4.045637215946841, 0.0], [0.0, 0.0, 4.045637215946841]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.02281860797342, 2.02281860797342], [2.02281860797342, 0.0, 2.02281860797342], [2.02281860797342, 2.02281860797342, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.9365262253598, 'volume': 66.21567448236252}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'd5fc7015-e7fe-4111-bf16-23bdc6755f93': {1: Response(output={'s_0': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.906019785962149, 0.0, 0.0], [0.0, 3.906019785962149, 0.0], [0.0, 0.0, 3.906019785962149]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.953009892981074, 1.953009892981074], [1.953009892981074, 0.0, 1.953009892981074], [1.953009892981074, 1.953009892981074, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 's_1': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.977053844318049, 0.0, 0.0], [0.0, 3.977053844318049, 0.0], [0.0, 0.0, 3.977053844318049]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.988526922159024, 1.988526922159024], [1.988526922159024, 0.0, 1.988526922159024], [1.988526922159024, 1.988526922159024, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 's_2': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.045637215946841, 0.0, 0.0], [0.0, 4.045637215946841, 0.0], [0.0, 0.0, 4.045637215946841]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.02281860797342, 2.02281860797342], [2.02281860797342, 0.0, 2.02281860797342], [2.02281860797342, 2.02281860797342, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 's_3': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.111970927282956, 0.0, 0.0], [0.0, 4.111970927282956, 0.0], [0.0, 0.0, 4.111970927282956]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.0559854636414774, 2.0559854636414774], [2.0559854636414774, 0.0, 2.0559854636414774], [2.0559854636414774, 2.0559854636414774, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 's_4': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.176230852372181, 0.0, 0.0], [0.0, 4.176230852372181, 0.0], [0.0, 0.0, 4.176230852372181]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.08811542618609, 2.08811542618609], [2.08811542618609, 0.0, 2.08811542618609], [2.08811542618609, 2.08811542618609, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}'}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'49b7b2e8-78ca-4d63-bfa6-f3b47ca3378f': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.977053844318049, 0.0, 0.0], [0.0, 3.977053844318049, 0.0], [0.0, 0.0, 3.977053844318049]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.988526922159024, 1.988526922159024], [1.988526922159024, 0.0, 1.988526922159024], [1.988526922159024, 1.988526922159024, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'scf', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'a45e0be5-c229-4817-8dc8-e2f54c46205f': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.045637215946841, 0.0, 0.0], [0.0, 4.045637215946841, 0.0], [0.0, 0.0, 4.045637215946841]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.02281860797342, 2.02281860797342], [2.02281860797342, 0.0, 2.02281860797342], [2.02281860797342, 2.02281860797342, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'scf', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'30bf90f1-57c2-474b-8bd2-d8a18061d85c': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.111970927282956, 0.0, 0.0], [0.0, 4.111970927282956, 0.0], [0.0, 0.0, 4.111970927282956]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.0559854636414774, 2.0559854636414774], [2.0559854636414774, 0.0, 2.0559854636414774], [2.0559854636414774, 2.0559854636414774, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'scf', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'd4b595f2-93a9-42d7-a547-f64be9b4ca80': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.176230852372181, 0.0, 0.0], [0.0, 4.176230852372181, 0.0], [0.0, 0.0, 4.176230852372181]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.08811542618609, 2.08811542618609], [2.08811542618609, 0.0, 2.08811542618609], [2.08811542618609, 2.08811542618609, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'scf', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'8c32a293-38d8-4c3a-82fc-78edd324d64d': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.906019785962149, 0.0, 0.0], [0.0, 3.906019785962149, 0.0], [0.0, 0.0, 3.906019785962149]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.953009892981074, 1.953009892981074], [1.953009892981074, 0.0, 1.953009892981074], [1.953009892981074, 1.953009892981074, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'pseudopotentials': {'Al': 'Al.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'kpts': [3, 3, 3], 'calculation': 'scf', 'smearing': 0.02}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'e747fb6a-e704-44ff-906e-664be75da6fc': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.9770538269346, 0.0, 0.0], [0.0, 3.9770538269346, 0.0], [0.0, 0.0, 3.9770538269346]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.9885269135082753, 1.9885269135082753], [1.9885269135082753, 0.0, 1.9885269135082753], [1.9885269135082753, 1.9885269135082753, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.9161488594614, 'volume': 62.904889933384574}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'579fbf80-e6e7-464c-a808-f5cecd9ef430': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.045637198263616, 0.0, 0.0], [0.0, 4.045637198263616, 0.0], [0.0, 0.0, 4.045637198263616]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.0228185991583882, 2.0228185991583882], [2.0228185991583882, 0.0, 2.0228185991583882], [2.0228185991583882, 2.0228185991583882, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.9365241668363, 'volume': 66.21567361408889}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'ceb70e3f-c90a-4aa3-a670-23ecf40fa11b': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.111970909309796, 0.0, 0.0], [0.0, 4.111970909309796, 0.0], [0.0, 0.0, 4.111970909309796]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.0559854546134178, 2.0559854546134178], [2.0559854546134178, 0.0, 2.0559854546134178], [2.0559854546134178, 2.0559854546134178, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.9192860025814, 'volume': 69.52645729479359}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'290a2a1d-9a74-40b1-af28-e4f61b357694': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[4.17623083411814, 0.0, 0.0], [0.0, 4.17623083411814, 0.0], [0.0, 0.0, 4.17623083411814]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 2.08811541707298, 2.08811541707298], [2.08811541707298, 0.0, 2.08811541707298], [2.08811541707298, 2.08811541707298, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.8737904693417, 'volume': 72.83724097549783}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'a9f810e8-c291-46b9-ab8f-4c59765bf3b9': {1: Response(output={'structure': '{"immutable_id": null, "last_modified": null, "elements": ["Al"], "nelements": 1, "elements_ratios": [1.0], "chemical_formula_descriptive": "Al4", "chemical_formula_reduced": "Al", "chemical_formula_hill": null, "chemical_formula_anonymous": "A", "dimension_types": [1, 1, 1], "nperiodic_dimensions": 3, "lattice_vectors": [[3.9060197688891853, 0.0, 0.0], [0.0, 3.9060197688891853, 0.0], [0.0, 0.0, 3.9060197688891853]], "space_group_symmetry_operations_xyz": null, "space_group_symbol_hall": null, "space_group_symbol_hermann_mauguin": null, "space_group_symbol_hermann_mauguin_extended": null, "space_group_it_number": null, "cartesian_site_positions": [[0.0, 0.0, 0.0], [0.0, 1.9530098844635175, 1.9530098844635175], [1.9530098844635175, 0.0, 1.9530098844635175], [1.9530098844635175, 1.9530098844635175, 0.0]], "nsites": 4, "species": [{"name": "Al", "chemical_symbols": ["Al"], "concentration": [1.0], "mass": null, "original_name": null, "attached": null, "nattached": null}], "species_at_sites": ["Al", "Al", "Al", "Al"], "assemblies": null, "structure_features": []}', 'energy': -1074.8457446150612, 'volume': 59.59410625268013}, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
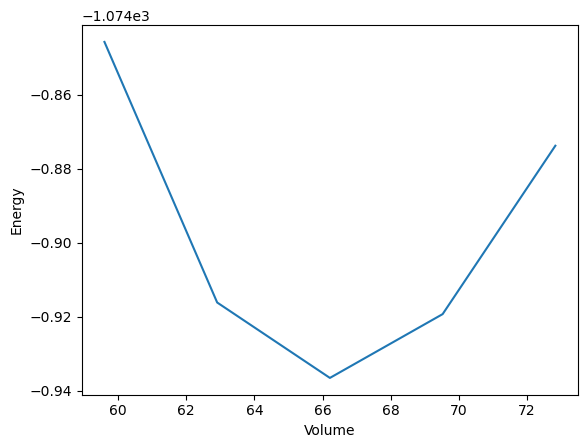
'20122c29-0e2d-4a9c-8506-d371390f8042': {1: Response(output=[-1074.8457446150612, -1074.9161488594614, -1074.9365241668363, -1074.9192860025814, -1074.8737904693417], detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'9842e2f0-3ca3-4a23-b82b-a3644a0294a8': {1: Response(output=[59.59410625268013, 62.904889933384574, 66.21567361408889, 69.52645729479359, 72.83724097549783], detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))},
'a1614162-0427-4cd2-a2c4-01837c5ae505': {1: Response(output=None, detour=None, addition=None, replace=None, stored_data=None, stop_children=False, stop_jobflow=False, job_dir=PosixPath('/home/jovyan/example_workflows/quantum_espresso'))}}

Load Workflow with pyiron_workflow#
from python_workflow_definition.pyiron_workflow import load_workflow_json
wf = load_workflow_json(file_name=workflow_json_filename)
wf.get_bulk_structure.inputs.a.value = 4.05
wf.draw(size=(10,10))
wf.run()
[jupyter-pythonworkflow-fl--x---218119f8:01978] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:01993] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02008] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02018] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02028] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02039] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
{'plot_energy_volume_curve__plot_energy_volume_curve': None}

