jobflow#
jobflow and atomate2 are key packages of the Materials Project . jobflow was especially designed to simplify the execution of dynamic workflows – when the actual number of jobs is dynamically determined upon runtime instead of being statically fixed before running the workflow(s). jobflow’s overall flexibility allows for building workflows that go beyond the usage in materials science. jobflow serves as the basis of atomate2, which implements data generation workflows in the context of materials science and will be used for data generation in the Materials Project in the future.
Define workflow with jobflow#
We start by importing the job decorator and Flow class from jobflow and the respective PWD tools.
import numpy as np
from jobflow import job, Flow
from python_workflow_definition.jobflow import write_workflow_json
Quantum Espresso Workflow#
We will use the knowledge from the previous arithmetic workflow example to create the Quantum Espresso-related tasks for calculating an “Energy vs. Volume” curve. It’s important to note that this is only a basic implementation, and further extensions towards data validation or for a simplified user experience can be added. For example, one can typically configure run commands for quantum-chemical programs via configuration files in atomate2.
from workflow import (
calculate_qe as _calculate_qe,
generate_structures as _generate_structures,
get_bulk_structure as _get_bulk_structure,
plot_energy_volume_curve as _plot_energy_volume_curve,
)
workflow_json_filename = "jobflow_qe.json"
calculate_qe = job(_calculate_qe)
generate_structures = job(_generate_structures)
plot_energy_volume_curve = job(_plot_energy_volume_curve)
get_bulk_structure = job(_get_bulk_structure)
We need to specify the typical QE input like pseudopotential(s) and structure model.
pseudopotentials = {"Al": "Al.pbe-n-kjpaw_psl.1.0.0.UPF"}
structure = get_bulk_structure(
element="Al",
a=4.04,
cubic=True,
)
calc_mini = calculate_qe(
working_directory="mini",
input_dict={
"structure": structure.output,
"pseudopotentials": pseudopotentials,
"kpts": (3, 3, 3),
"calculation": "vc-relax",
"smearing": 0.02,
},
)
Next, for the “Energy vs. Volume” curve, we meed to specify the number of strained structures and save them into a list object. For each of the strained structures, we will carry out a QE calculation.
number_of_strains = 5
structure_lst = generate_structures(
structure=calc_mini.output.structure,
strain_lst=np.linspace(0.9, 1.1, number_of_strains),
)
job_strain_lst = []
for i in range(number_of_strains):
calc_strain = calculate_qe(
working_directory="strain_" + str(i),
input_dict={
"structure": getattr(structure_lst.output, f"s_{i}"),
"pseudopotentials": pseudopotentials,
"kpts": (3, 3, 3),
"calculation": "scf",
"smearing": 0.02,
},
)
job_strain_lst.append(calc_strain)
Finally, we specify a plotter for the “Energy vs. Volume” curve and can export the workflow.
plot = plot_energy_volume_curve(
volume_lst=[job.output.volume for job in job_strain_lst],
energy_lst=[job.output.energy for job in job_strain_lst],
)
flow = Flow([structure, calc_mini, structure_lst] + job_strain_lst + [plot])
write_workflow_json(flow=flow, file_name=workflow_json_filename)
!cat {workflow_json_filename}
{
"version": "0.1.0",
"nodes": [
{
"id": 0,
"type": "function",
"value": "workflow.get_bulk_structure"
},
{
"id": 1,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 2,
"type": "function",
"value": "workflow.generate_structures"
},
{
"id": 3,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 4,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 5,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 6,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 7,
"type": "function",
"value": "workflow.calculate_qe"
},
{
"id": 8,
"type": "function",
"value": "workflow.plot_energy_volume_curve"
},
{
"id": 9,
"type": "input",
"name": "element",
"value": "Al"
},
{
"id": 10,
"type": "input",
"name": "a",
"value": 4.04
},
{
"id": 11,
"type": "input",
"name": "cubic",
"value": true
},
{
"id": 12,
"type": "input",
"name": "working_directory_0",
"value": "mini"
},
{
"id": 13,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 14,
"type": "input",
"name": "pseudopotentials",
"value": {
"Al": "Al.pbe-n-kjpaw_psl.1.0.0.UPF"
}
},
{
"id": 15,
"type": "input",
"name": "kpts",
"value": [
3,
3,
3
]
},
{
"id": 16,
"type": "input",
"name": "calculation_0",
"value": "vc-relax"
},
{
"id": 17,
"type": "input",
"name": "smearing",
"value": 0.02
},
{
"id": 18,
"type": "input",
"name": "strain_lst",
"value": [
0.9,
0.9500000000000001,
1.0,
1.05,
1.1
]
},
{
"id": 19,
"type": "input",
"name": "working_directory_1",
"value": "strain_0"
},
{
"id": 20,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 21,
"type": "input",
"name": "calculation_1",
"value": "scf"
},
{
"id": 22,
"type": "input",
"name": "working_directory_2",
"value": "strain_1"
},
{
"id": 23,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 24,
"type": "input",
"name": "working_directory_3",
"value": "strain_2"
},
{
"id": 25,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 26,
"type": "input",
"name": "working_directory_4",
"value": "strain_3"
},
{
"id": 27,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 28,
"type": "input",
"name": "working_directory_5",
"value": "strain_4"
},
{
"id": 29,
"type": "function",
"value": "python_workflow_definition.shared.get_dict"
},
{
"id": 30,
"type": "function",
"value": "python_workflow_definition.shared.get_list"
},
{
"id": 31,
"type": "function",
"value": "python_workflow_definition.shared.get_list"
},
{
"id": 32,
"type": "output",
"name": "result"
}
],
"edges": [
{
"target": 0,
"targetPort": "element",
"source": 9,
"sourcePort": null
},
{
"target": 0,
"targetPort": "a",
"source": 10,
"sourcePort": null
},
{
"target": 0,
"targetPort": "cubic",
"source": 11,
"sourcePort": null
},
{
"target": 1,
"targetPort": "working_directory",
"source": 12,
"sourcePort": null
},
{
"target": 13,
"targetPort": "structure",
"source": 0,
"sourcePort": null
},
{
"target": 13,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 13,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 13,
"targetPort": "calculation",
"source": 16,
"sourcePort": null
},
{
"target": 13,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 1,
"targetPort": "input_dict",
"source": 13,
"sourcePort": null
},
{
"target": 2,
"targetPort": "structure",
"source": 1,
"sourcePort": "structure"
},
{
"target": 2,
"targetPort": "strain_lst",
"source": 18,
"sourcePort": null
},
{
"target": 3,
"targetPort": "working_directory",
"source": 19,
"sourcePort": null
},
{
"target": 20,
"targetPort": "structure",
"source": 2,
"sourcePort": "s_0"
},
{
"target": 20,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 20,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 20,
"targetPort": "calculation",
"source": 21,
"sourcePort": null
},
{
"target": 20,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 3,
"targetPort": "input_dict",
"source": 20,
"sourcePort": null
},
{
"target": 4,
"targetPort": "working_directory",
"source": 22,
"sourcePort": null
},
{
"target": 23,
"targetPort": "structure",
"source": 2,
"sourcePort": "s_1"
},
{
"target": 23,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 23,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 23,
"targetPort": "calculation",
"source": 21,
"sourcePort": null
},
{
"target": 23,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 4,
"targetPort": "input_dict",
"source": 23,
"sourcePort": null
},
{
"target": 5,
"targetPort": "working_directory",
"source": 24,
"sourcePort": null
},
{
"target": 25,
"targetPort": "structure",
"source": 2,
"sourcePort": "s_2"
},
{
"target": 25,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 25,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 25,
"targetPort": "calculation",
"source": 21,
"sourcePort": null
},
{
"target": 25,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 5,
"targetPort": "input_dict",
"source": 25,
"sourcePort": null
},
{
"target": 6,
"targetPort": "working_directory",
"source": 26,
"sourcePort": null
},
{
"target": 27,
"targetPort": "structure",
"source": 2,
"sourcePort": "s_3"
},
{
"target": 27,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 27,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 27,
"targetPort": "calculation",
"source": 21,
"sourcePort": null
},
{
"target": 27,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 6,
"targetPort": "input_dict",
"source": 27,
"sourcePort": null
},
{
"target": 7,
"targetPort": "working_directory",
"source": 28,
"sourcePort": null
},
{
"target": 29,
"targetPort": "structure",
"source": 2,
"sourcePort": "s_4"
},
{
"target": 29,
"targetPort": "pseudopotentials",
"source": 14,
"sourcePort": null
},
{
"target": 29,
"targetPort": "kpts",
"source": 15,
"sourcePort": null
},
{
"target": 29,
"targetPort": "calculation",
"source": 21,
"sourcePort": null
},
{
"target": 29,
"targetPort": "smearing",
"source": 17,
"sourcePort": null
},
{
"target": 7,
"targetPort": "input_dict",
"source": 29,
"sourcePort": null
},
{
"target": 30,
"targetPort": "0",
"source": 3,
"sourcePort": "volume"
},
{
"target": 30,
"targetPort": "1",
"source": 4,
"sourcePort": "volume"
},
{
"target": 30,
"targetPort": "2",
"source": 5,
"sourcePort": "volume"
},
{
"target": 30,
"targetPort": "3",
"source": 6,
"sourcePort": "volume"
},
{
"target": 30,
"targetPort": "4",
"source": 7,
"sourcePort": "volume"
},
{
"target": 8,
"targetPort": "volume_lst",
"source": 30,
"sourcePort": null
},
{
"target": 31,
"targetPort": "0",
"source": 3,
"sourcePort": "energy"
},
{
"target": 31,
"targetPort": "1",
"source": 4,
"sourcePort": "energy"
},
{
"target": 31,
"targetPort": "2",
"source": 5,
"sourcePort": "energy"
},
{
"target": 31,
"targetPort": "3",
"source": 6,
"sourcePort": "energy"
},
{
"target": 31,
"targetPort": "4",
"source": 7,
"sourcePort": "energy"
},
{
"target": 8,
"targetPort": "energy_lst",
"source": 31,
"sourcePort": null
},
{
"target": 32,
"targetPort": null,
"source": 8,
"sourcePort": null
}
]
}
Load Workflow with aiida#
Now, we can import the workflow, run it with aiida and plot the “Energy vs. Volume” curve.
from aiida import orm, load_profile
load_profile()
Profile<uuid='1682a1dbda304ec998030372ef5350ab' name='pwd'>
from python_workflow_definition.aiida import load_workflow_json
wg = load_workflow_json(workflow_json_filename)
wg.nodes.get_bulk_structure.inputs.a.value = orm.Float(4.05)
wg
wg.run()
05/26/2025 04:59:05 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_bulk_structure1
05/26/2025 04:59:05 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_bulk_structure1, type: PyFunction, finished.
05/26/2025 04:59:05 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict10
05/26/2025 04:59:06 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict10, type: PyFunction, finished.
05/26/2025 04:59:06 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe2
[jupyter-pythonworkflow-fl--x---218119f8:02569] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 04:59:56 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['energy', 'volume'])
05/26/2025 04:59:57 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe2, type: PyFunction, finished.
05/26/2025 04:59:57 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: generate_structures3
05/26/2025 04:59:58 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: generate_structures3, type: PyFunction, finished.
05/26/2025 04:59:58 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict11,get_dict12,get_dict13,get_dict14,get_dict15
05/26/2025 04:59:58 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict11, type: PyFunction, finished.
05/26/2025 04:59:58 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe4,get_dict12,get_dict13,get_dict14,get_dict15
[jupyter-pythonworkflow-fl--x---218119f8:02586] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 05:00:10 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 05:00:10 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe4, type: PyFunction, finished.
05/26/2025 05:00:10 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict12,get_dict13,get_dict14,get_dict15
05/26/2025 05:00:11 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict12, type: PyFunction, finished.
05/26/2025 05:00:11 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe5,get_dict13,get_dict14,get_dict15
[jupyter-pythonworkflow-fl--x---218119f8:02596] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 05:00:22 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 05:00:22 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe5, type: PyFunction, finished.
05/26/2025 05:00:22 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict13,get_dict14,get_dict15
05/26/2025 05:00:23 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict13, type: PyFunction, finished.
05/26/2025 05:00:23 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe6,get_dict14,get_dict15
[jupyter-pythonworkflow-fl--x---218119f8:02606] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 05:00:35 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 05:00:35 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe6, type: PyFunction, finished.
05/26/2025 05:00:35 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict14,get_dict15
05/26/2025 05:00:35 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict14, type: PyFunction, finished.
05/26/2025 05:00:36 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe7,get_dict15
[jupyter-pythonworkflow-fl--x---218119f8:02616] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 05:00:50 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 05:00:51 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe7, type: PyFunction, finished.
05/26/2025 05:00:51 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_dict15
05/26/2025 05:00:51 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_dict15, type: PyFunction, finished.
05/26/2025 05:00:51 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: calculate_qe8
[jupyter-pythonworkflow-fl--x---218119f8:02626] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
05/26/2025 05:01:05 AM <2497> aiida.orm.nodes.process.calculation.calcfunction.CalcFunctionNode: [WARNING] Found extra results that are not included in the output: dict_keys(['structure'])
05/26/2025 05:01:05 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: calculate_qe8, type: PyFunction, finished.
05/26/2025 05:01:06 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_list16,get_list17
05/26/2025 05:01:06 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_list16, type: PyFunction, finished.
05/26/2025 05:01:07 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: get_list17
05/26/2025 05:01:07 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: get_list17, type: PyFunction, finished.
05/26/2025 05:01:07 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run: plot_energy_volume_curve9
05/26/2025 05:01:08 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|update_task_state]: Task: plot_energy_volume_curve9, type: PyFunction, finished.
05/26/2025 05:01:08 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|continue_workgraph]: tasks ready to run:
05/26/2025 05:01:08 AM <2497> aiida.orm.nodes.process.workflow.workchain.WorkChainNode: [REPORT] [111|WorkGraphEngine|finalize]: Finalize workgraph.
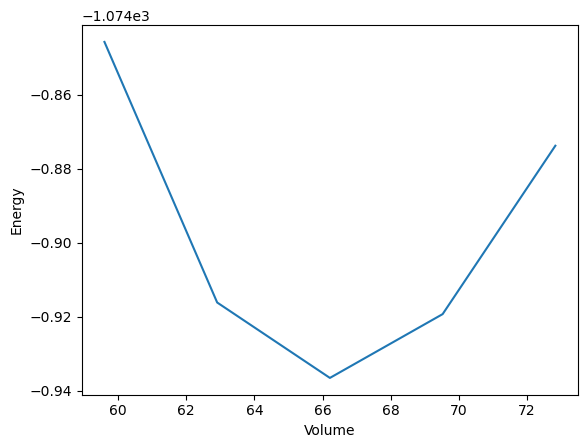
Load Workflow with pyiron_base#
And we can repeat the same process using pyiron.
from python_workflow_definition.pyiron_base import load_workflow_json
delayed_object_lst = load_workflow_json(file_name=workflow_json_filename)
delayed_object_lst[-1].draw()
delayed_object_lst[0].input['a'] = 4.05
delayed_object_lst[-1].pull()
The job get_bulk_structure_2ca4aeae204ceaa28593c93054b07908 was saved and received the ID: 1
The job get_dict_1e47509b88d63a21fd421686554c8f4a was saved and received the ID: 2
The job calculate_qe_411e578f2700d09ba2df9a4c682b4582 was saved and received the ID: 3
[jupyter-pythonworkflow-fl--x---218119f8:02656] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job generate_structures_550221ce8aa27fbdf50dbf13e059ce24 was saved and received the ID: 4
The job get_dict_68a4c6ff00bffbd8f249d6022ec8abb2 was saved and received the ID: 5
The job calculate_qe_8e11594357cf470d576634593684ae66 was saved and received the ID: 6
[jupyter-pythonworkflow-fl--x---218119f8:02670] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job get_dict_64aacd2badb7b0a126a5662dabd1ce0c was saved and received the ID: 7
The job calculate_qe_95276d45f24b5e6e645aab15a3b853df was saved and received the ID: 8
[jupyter-pythonworkflow-fl--x---218119f8:02680] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job get_dict_7079cab3db71ad163cfc2e39fcf98688 was saved and received the ID: 9
The job calculate_qe_3c1dbc8b42e651e328ae99033a426980 was saved and received the ID: 10
[jupyter-pythonworkflow-fl--x---218119f8:02690] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job get_dict_0e89a9ec0ef1fd918240aa8d2dc44178 was saved and received the ID: 11
The job calculate_qe_2550d6d1ccd7ab3d68c384d3887a3fe3 was saved and received the ID: 12
[jupyter-pythonworkflow-fl--x---218119f8:02700] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job get_dict_2ac7f25535cfa6c6850c0f9af197b541 was saved and received the ID: 13
The job calculate_qe_10d82eb36a247279f62c63b70e1a55d2 was saved and received the ID: 14
[jupyter-pythonworkflow-fl--x---218119f8:02710] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
The job get_list_e21c87f95f3795055e7214eec6d7c927 was saved and received the ID: 15
The job get_list_61df540ee8a34a06782249cbe8834865 was saved and received the ID: 16
The job plot_energy_volume_curve_bbe171c30a8de804ba8d4227e65e62bf was saved and received the ID: 17

Load Workflow with pyiron_workflow#
from python_workflow_definition.pyiron_workflow import load_workflow_json
wf = load_workflow_json(file_name=workflow_json_filename)
wf.get_bulk_structure.inputs.a.value = 4.05
wf.draw(size=(10,10))
wf.run()
[jupyter-pythonworkflow-fl--x---218119f8:02722] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02738] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02748] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02758] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02768] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
[jupyter-pythonworkflow-fl--x---218119f8:02778] mca_base_component_repository_open: unable to open mca_btl_openib: librdmacm.so.1: cannot open shared object file: No such file or directory (ignored)
Note: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG
{'plot_energy_volume_curve__plot_energy_volume_curve': None}

